Lab Notebook: Estimating body height in ant locomotion
- Description: Import, process and visualize body height of two different ant species
- Keywords: Cataglyphis, Formica, body height, center of mass, inclined locomotion, ant
- Requirements: norm_t.m impHedrickLab.m
- Disclaimer: Without warranty of any kind. Content may change.
- Language: english, matlab
- Author: T. Wöhrl
- URL: https://t-woehrl.net/p/20191105T212420/index.html
- License: MIT
Contents
TODO
- height over time (oscillation, undulation)
Changelog
- 2019-11-07: Requirements, compare results with literature results
- 2019-11-06: Published note
- 2019-11-05: Created note
Initialize
clear all; close all; clc version
ans = 9.1.0.441655 (R2016b)
Import meta data
p.meta.filename = ['G:/dropbox/todo/ant_impulse/dat/ant_metadata.xlsx']; p.meta.sheetname = 'Protokoll'; a = exist('exl'); if a == 0 exl = dataset('XLSFile',p.meta.filename,... 'Sheet','Protokoll','range','A5:DZ2425'); end
Subsetting
select tracked data (criteria: no stopping, no starting, gait cycle within capture window)
fil = (exl.W2014force == 1 ) ;
% fil = (exl.W2014force == 1 & (char(exl.ant) == 'C') ) ;
sub1 = exl(fil,exl.Properties.VarNames);
n_sub1 = length(sub1);
Import data and calculate
% Kinematic data root p.raw.kin = 'G:/archive/dat/120901_digitalization/'; % data einheit.kin = 10.^(-3); % units % Error handling err = []; % results dataset d = []; for k = 1:n_sub1 % display progress % disp([num2str(k),'/',num2str(n_sub1),'/',num2str(sum(err))]); % error handling try % Import kinematic data id = char(sub1.id(k)); datadir = [p.raw.kin,id]; % import center of origin clear xy0 xz0 n xy0 = impHedrickLab(datadir,id,'x0','txt'); xz0 = impHedrickLab(datadir,id,'z0','txt'); % number of data points n.kin = length(xy0); % only first row cotains relevant data xy0 = repmat(xy0(1,:),n.kin,1); xz0 = repmat(xz0(1,:),n.kin,1); % import petiolous marker b3 = impHedrickLab(datadir,id,'b3','txt') - xy0; b3z = impHedrickLab(datadir,id,'b3z','txt')- xz0; % average for x value, x in two planes b3(:,1) = mean([b3(:,1) b3z(:,1)],2); % import neck marker b2 = impHedrickLab(datadir,id,'b2','txt') - xy0; b2z = impHedrickLab(datadir,id,'b2z','txt')- xz0; % average for x value, x in two planes b2(:,1) = mean([b2(:,1) b2z(:,1)],2); % select data for one gait cycle (touchdown leg 2) cut.Gait = [sub1.td12(k):1:sub1.td22(k)]; b3 = b3(cut.Gait,:); b3(:,3) = b3z(cut.Gait,2); b2 = b2(cut.Gait,:); b2(:,3) = b2z(cut.Gait,2); % estimate center of mass location b2b3(k,1) = mean(norm_t(b2-b3)); if strcmp(sub1.ant(k),'C')
for Cataglyphis: "This distance represents 5% (20%) of the length of the thorax." McMeeking, R. M.; Arzt, E. & Wehner, R. Cataglyphis desert ants improve their mobility by raising the gaster J. Theoret. Biol., 2012, 297, 17-25
COM = b3 + (b2-b3)./repmat(norm_t(b2-b3),1,3).*b2b3(k,1).*0.2;
elseif strcmp(sub1.ant(k),'F')
for Formica: "Marker B3 was located on average less than 0.3 mm away from the CoM." Reinhardt, L. & Blickhan, R. Level locomotion in wood ants: evidence for grounded running J. Exp. Biol., 2014, 217, 2358-2370 COM = b3;
COM = b3 + (b2-b3)./repmat(norm_t(b2-b3),1,3).*0.3;
end % allocate data d.COMheight(k,1) = mean(COM(:,3)).*einheit.kin; d.Petiolousheight(k,1) = mean(b3(:,3)).*einheit.kin ; d.Neckheight(k,1) = mean(b2(:,3)).*einheit.kin; d.FramesKin(k,1) = n.kin; d.ant(k,1) = sub1.ant(k); d.m(k,1) = sub1.m(k); d.deg(k,1) = sub1.deg(k); d.id{k,1} = id; d.err(k,1) = 0; catch me err(k) = 1; d.err(k,1) = 1; end end
Errors
sub1.id(logical(err)); d.ant(isnan(d.COMheight));
Table summary
grpstats(dataset(d),{'ant','deg'},{'nanmean','nanmedian','nanstd'},...
'DataVars','COMheight')
ans =
ant deg GroupCount nanmean_COMheight
C_-60 'C' -60 45 0.0025814
C_-30 'C' -30 45 0.0036085
C_0 'C' 0 45 0.0038155
C_30 'C' 30 45 0.0030433
C_60 'C' 60 45 0.0025091
F_-90 'F' -90 42 0.0025737
F_-60 'F' -60 45 0.0022442
F_-30 'F' -30 45 0.0024738
F_0 'F' 0 45 0.0025671
F_30 'F' 30 45 0.0021281
F_60 'F' 60 45 0.0024673
F_90 'F' 90 15 0.0022549
nanmedian_COMheight nanstd_COMheight
C_-60 0.0023784 0.00065322
C_-30 0.003518 0.00043904
C_0 0.0038414 0.00068379
C_30 0.0030213 0.00043018
C_60 0.0024237 0.00047596
F_-90 0.0025883 0.00031913
F_-60 0.0021919 0.00024084
F_-30 0.0024724 0.00028236
F_0 0.0025029 0.00054191
F_30 0.0020584 0.00031102
F_60 0.0025194 0.00030705
F_90 0.0023294 0.00028427
Plot boxplot
bh = boxplot(d.COMheight./einheit.kin,{d.ant,d.deg},'factorseparator',[1 1],...
'labelverbosity','majorminor','medianstyle','line','notch','on',...
'colorgroup',{d.ant},'boxstyle',...
'outline','factordirection','list','boxstyle','outline',...
'symbol','+k','jitter',0.5);
title('Draft for: Slope-dependent body height adjustments in ant locomotion')
xlabel({'Species';'Slope in deg'})
ylabel('COMheight in mm')
shg
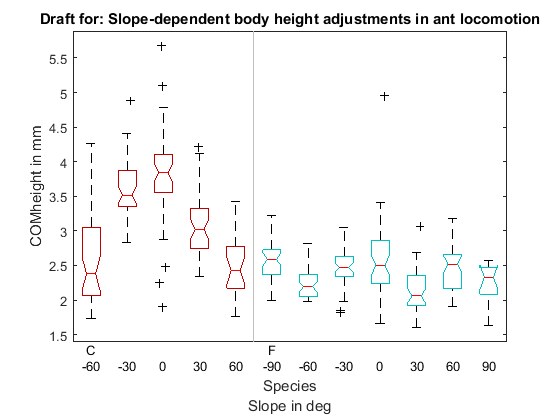
From: Reinhardt, L. & Blickhan, R. Level locomotion in wood ants: evidence for grounded running J. Exp. Biol., 2014, 217, 2358-2370
- Formica polyctena: Center of mass height 2.43±0.39 mm for speed v < 11.5 cm s^(-1) and 2.62±0.38 mm for speed v > 11.5 cm s^(-1)
From: Weihmann, T. & Blickhan, R. Comparing inclined locomotion in a ground-living and a climbing ant species: sagittal plane kinematics. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol., 2009, 195, 1011-1020
- Formica pratensis: Center of mass height: 0.59 altitrunk length of 3.15±0.25 mm for level locomotion
- Cataglyphis fortis: 0.74 altitrunk length of 3.73±0.24 mm for level locomotion