Code for: Propulsion in hexapod locomotion: How do desert ants traverse slopes?
Contents
- Bibliographic Information
- Example Video
- Prerequisites
- Load data
- Fig. 1A-C: Weight-specific leg impulses at different slopes
- Fig. 2A-E: Mean GRF vectors with mean stepping patterns at different slopes
- Fig. 3A-C: GRF over time
- Fig. 4A-D: Comparison of the mean GRF vectors on upslopes and downslopes
- Fig. 5A-B: Hind leg dragging and change of body height
- Fig. 5A: Hind leg dragging
- Fig. 5B: Change of body height
- Fig. 6A-C: Normalised double support durations for each leg pair at different slopes
- Table 2: Dragging hind leg measurements
- Table 4: Kinematic parameter
- Table 4: Body Height (Center of mass - substrate distance in mm)
- Body Height (Petiole - substrate distance in mm)
Bibliographic Information
% Manuscript Decision - Accepted on 7 Feb 2017 % MS ID#: JEXBIO/2016/137505 % MS TITLE: Propulsion in hexapod locomotion: How do desert ants traverse slopes? % AUTHORS: Toni Woehrl, Lars Reinhardt, and Reinhard Blickhan % ARTICLE TYPE: Research Article % Journal: Journal of Experimental Biology 2017 220: 1618-1625 % DOI (Data): 10.5061/dryad.j4594, published online February 22, 2017 % DOI (Article): 10.1242/jeb.137505, published online May 3, 2017
Example Video
Please click on the image below to download the video.
Prerequisites
The following two additional Matlab scripts were required to plot the figures.
- stdshade.m 2010-12-01 Source: http://www.mathworks.com/matlabcentral/fileexchange/29534-stdshade
- tight_subplot.m 2010-06-20 Source: http://www.mathworks.com/matlabcentral/fileexchange/27991-tight-subplot-nh--nw--gap--marg-h--marg-w-
Matlab version
version
ans =
8.6.0.267246 (R2015b)
Load data
Download: data.mat (5.48 MB)
close all; clear all; clc load('data.mat') whos
Name Size Bytes Class Attributes F 6-D 2160000 double d3 75x4 4594 dataset data1 225x19 138924 dataset data2 1x1 5138670 struct p5l 1x1 120352 struct
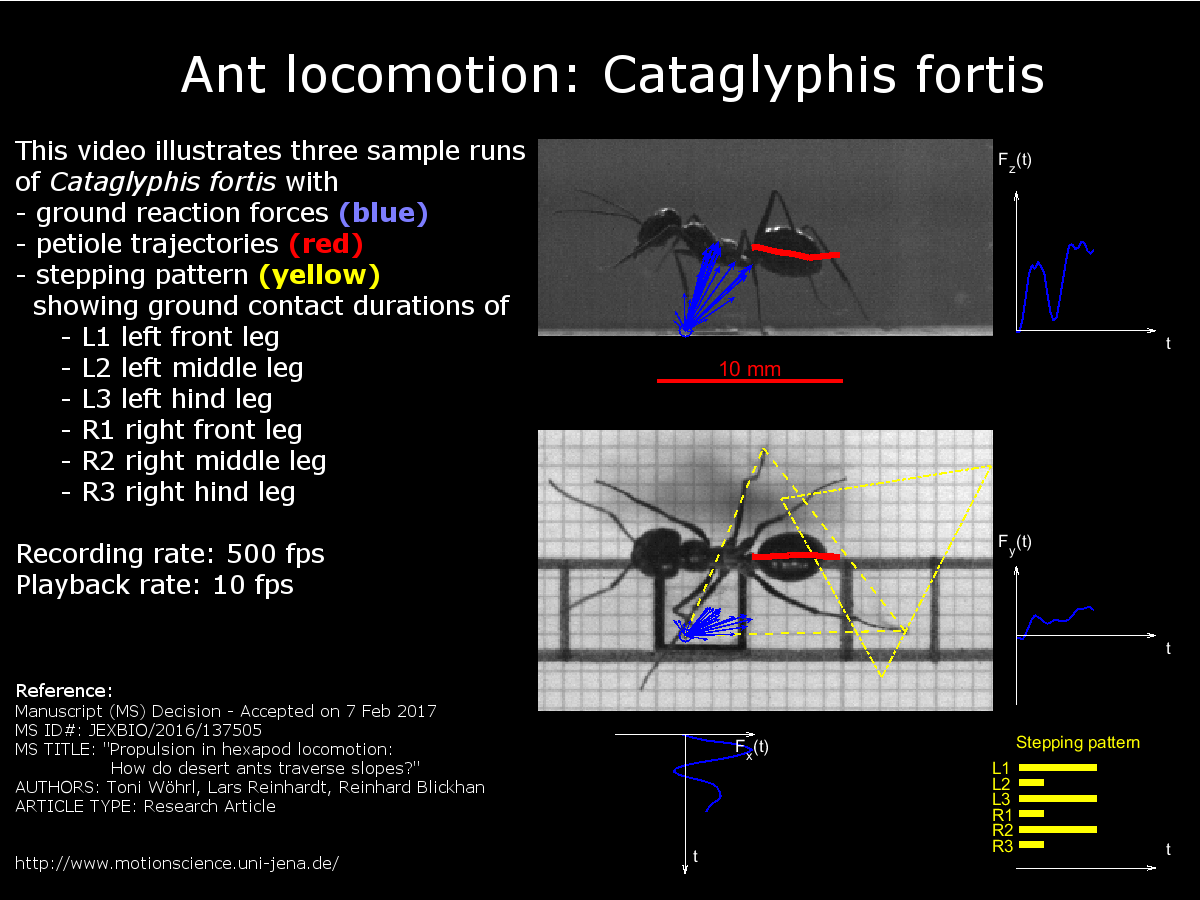
Fig. 1A-C: Weight-specific leg impulses at different slopes
plot_data1.title = {'Fore-aft Impulse J_x', 'Lateral Impulse J_y', 'Normal Impulse J_z'};
plot_data1.color = [1 0 0 ; 0 0 1 ; 0 0.7 0];
plot_data1.direction = 'xyz';
leg = unique(data1.legnr);
slope = unique(data1.deg);
% location of horizontal lines below siginificance stars for
% pairwise comparisons
y1 = [45 45];
y2 = [40 40]; % neighbours
xd = 0.05; % gap
% Plot
for l = 1:length(plot_data1.direction) % Direction
% Impulse data
clear I_l
I_l = eval(['data1.Impulse',plot_data1.direction(l),'BW']);
figure(l);
plot([0 16],[0 0],':k','LineWidth',0.5); hold on % Zero
% Plot data points (grey) below boxplots
for ll = 1:length(leg) % leg number
for lll = 1:length(slope) % slope
% subset impulse data
clear filter_lll I
filter_lll = logical(cell2mat(data1.legnr) == cell2mat(leg(ll)) & data1.deg == slope(lll));
% Impulse
I = I_l(filter_lll);
% scatter points horizontally
clear x lx x2
x = (ll-1)*length(slope) + lll;
lx = length(I);
x2 = linspace(-.1,.1,lx);
plot(x2+x,I,'.','Color',[1 1 1]*.7); hold all
% horizontal lines for significance stars
if lll==1 || lll==3
plot([x+xd x+2-xd],y1,'k')
end
if lll<5
plot([x+xd x+1-xd],y2,'k');
end
end
end
% plot boxplot
clear bh
bh = boxplot(I_l,{str2num(cell2mat(data1.legnr)),data1.deg},...
'factorseparator',[1 1],...
'labelverbosity','majorminor',...
'medianstyle','line',...
'notch','off',...
'colorgroup',data1.legnr,...
'color',plot_data1.color,...
'factordirection','list',...
'boxstyle','outline',...
'outliersize',0.1,...
'widths',.75);
set(bh, 'markeredgecolor', [1 1 1]*.5,'Linewidth',.5, 'linestyle','-');
axis([0.5 15.5 -50 50]);
axis square
title(plot_data1.title{l});
xlabel('Slope i (deg)');
ylabel('Weight-specific Impulse J_{lk} (ms)');
text(1,-42, {'Left';'front leg'} ,'color','k','horizontalalignment','left');
text(6,-42,{'Right';'middle leg'},'color','k','horizontalalignment','left');
text(11,-42,{'Left';'hind leg'} ,'color','k','horizontalalignment','left');
end
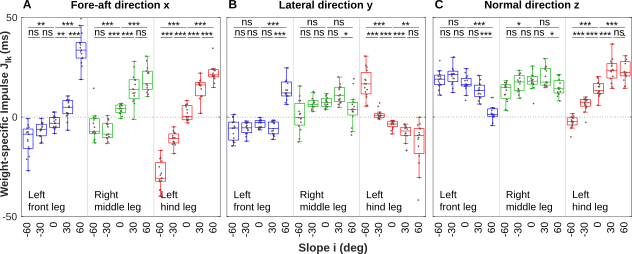
Fig. 2A-E: Mean GRF vectors with mean stepping patterns at different slopes
close all; % select and order slopes slopes = {'m60','m30','n00','p30','p60'}; % color coding of legs cl_kk = [0 0 1; 0 .7 0; 1 0 0]; % color and linestyle supporting tripd cl_sm = [1 1 1].*0; ls_sm = '--'; for k = 1:length(slopes) % Suporting tripod: tarsus midstance positions, suporting tripod (mean) clear msx1 msy1 msx2 msy2 msx3 msy3 msx1 = mean(data2.C.(slopes{k}).l1.MidStance2X,2); msy1 = mean(data2.C.(slopes{k}).l1.MidStance2Y,2); msx2 = mean(data2.C.(slopes{k}).l2.MidStance2X,2); msy2 = - mean(data2.C.(slopes{k}).l2.MidStance2Y,2); % middle leg lateral direction msx3 = mean(data2.C.(slopes{k}).l3.MidStance2X,2); msy3 = mean(data2.C.(slopes{k}).l3.MidStance2Y,2); % Displacement of petiolus (mean) clear dp dp = mean([data2.C.(slopes{k}).l1.Displacement,... data2.C.(slopes{k}).l2.Displacement,... data2.C.(slopes{k}).l3.Displacement],2); % Petiolus b3 clear b3px b3py b3pz b3px = [data2.C.(slopes{k}).l1.b3x,data2.C.(slopes{k}).l2.b3x,data2.C.(slopes{k}).l3.b3x]; b3py = [data2.C.(slopes{k}).l1.b3y,data2.C.(slopes{k}).l2.b3y,data2.C.(slopes{k}).l3.b3y]; b3pz = [data2.C.(slopes{k}).l1.b3z,data2.C.(slopes{k}).l2.b3z,data2.C.(slopes{k}).l3.b3z]; % Head-Thorax joint b2 clear b2px b2py b2pz b2px = [data2.C.(slopes{k}).l1.b2x,data2.C.(slopes{k}).l2.b2x,data2.C.(slopes{k}).l3.b2x]; b2py = [data2.C.(slopes{k}).l1.b2y,data2.C.(slopes{k}).l2.b2y,data2.C.(slopes{k}).l3.b2y]; b2pz = [data2.C.(slopes{k}).l1.b2z,data2.C.(slopes{k}).l2.b2z,data2.C.(slopes{k}).l3.b2z]; % Center of mass (mean): 20 % of the thorax length % McMeeking, R. M.; Arzt, E. and Wehner, R. (2012). Cataglyphis desert ants improve their mobility by raising the gaster. J. Theoret. Biol. 297, 17-25. clear compx compy compz compx = [data2.C.(slopes{k}).l1.comx,data2.C.(slopes{k}).l2.comx,data2.C.(slopes{k}).l3.comx]; compy = [data2.C.(slopes{k}).l1.comy,data2.C.(slopes{k}).l2.comy,data2.C.(slopes{k}).l3.comy]; compz = [data2.C.(slopes{k}).l1.comz,data2.C.(slopes{k}).l2.comz,data2.C.(slopes{k}).l3.comz]; % Mean body posture per slope and stride cycles, relative to Center of mass (x=0,y=0) b2px = mean(mean(b2px - compx,2)); b2py = mean(mean(b2py - compy,2)); b2pz = mean(mean(b2pz,2)); b3px = mean(mean(b3px - compx,2)); b3py = mean(mean(b3py - compy,2)); b3pz = mean(mean(b3pz,2)); for kk = 1:3 % legs hold all % Ground reaction force (duration,direction) clear GRF GRF(:,1) = mean(data2.C.(slopes{k}).(['l',num2str(kk)]).Fx,2); GRF(:,2) = mean(data2.C.(slopes{k}).(['l',num2str(kk)]).Fy,2); GRF(:,3) = mean(data2.C.(slopes{k}).(['l',num2str(kk)]).Fz,2); % Petiole (duration,direction) clear b3x b3y b3z b3x = data2.C.(slopes{k}).(['l',num2str(kk)]).b3x; b3y = data2.C.(slopes{k}).(['l',num2str(kk)]).b3y; b3z = data2.C.(slopes{k}).(['l',num2str(kk)]).b3z; % Head Thorax joint (duration, direction) clear b2x b2y b2z b2x = data2.C.(slopes{k}).(['l',num2str(kk)]).b2x; b2y = data2.C.(slopes{k}).(['l',num2str(kk)]).b2y; b2z = data2.C.(slopes{k}).(['l',num2str(kk)]).b2z; % Center of mass (duration, direction) clear comx comy comz comx = data2.C.(slopes{k}).(['l',num2str(kk)]).comx; comy = data2.C.(slopes{k}).(['l',num2str(kk)]).comy; comz = data2.C.(slopes{k}).(['l',num2str(kk)]).comz; % Tarsi (duration, direction) clear p5x p5y p5x = data2.C.(slopes{k}).(['l',num2str(kk)]).p5x; p5y = data2.C.(slopes{k}).(['l',num2str(kk)]).p5y; % Mean Body Height clear BodyHeight BodyHeight = data2.C.(slopes{k}).(['l',num2str(kk)]).BodyHeight; % mirror middle leg in lateral direction if kk == 2 p5y = -p5y; b3y = -b3y; b2y = -b2y; comy= -comy; GRF(:,2) = -GRF(:,2); end figure(k) %%%%%%%%%%%%%%%%%%%%%%%%% % Dorsal view (xy plane)% %%%%%%%%%%%%%%%%%%%%%%%%% subplot(2,1,2) % plot tarsus trajectories with mean clear px py pxm pym px = p5x - comx; py = p5y - comy; pxm = mean(p5x-comx,2); pym = mean(p5y-comy,2); plot(px,py,'color',[1 1 1].*.6); hold on plot(pxm,pym,'k','linewidth',2); % Supporting tripod (spatial stepping pattern) % plot layer over tarsus trajectories if kk == 3 % Tripod 1 plot([msx1 msx2],[msy1 msy2],'color',cl_sm,'linestyle',ls_sm); hold on plot([msx1 msx3],[msy1 msy3],'color',cl_sm,'linestyle',ls_sm) plot([msx2 msx3],[msy2 msy3],'color',cl_sm,'linestyle',ls_sm) plot(dp+[msx1 msx2],[msy1 msy2],'color',cl_sm,'linestyle',ls_sm); hold on plot(dp+[msx1 msx3],[msy1 msy3],'color',cl_sm,'linestyle',ls_sm) plot(dp+[msx2 msx3],[msy2 msy3],'color',cl_sm,'linestyle',ls_sm) % Tripod 2 plot(dp/2+[msx1 msx2],-[msy1 msy2],'color',[1 1 1].*.0,'linestyle',':'); hold on plot(dp/2+[msx1 msx3],-[msy1 msy3],'color',[1 1 1].*.0,'linestyle',':') plot(dp/2+[msx2 msx3],-[msy2 msy3],'color',[1 1 1].*.0,'linestyle',':') % Plot Thorax (Petiole -> Head-Thorax-joint) plot([b3px b2px],[b3py b2py],'color',[1 .5 0]*1,'linewidth',2) % Plot Displacement plot(0,0,'o','MarkerFaceColor',[1 .5 0]*1,... 'MarkerEdgeColor',[1 .5 0]*1,'Markersize',4); plot(0,0,'*k','Markersize',3); plot(dp,0,'o','MarkerFaceColor',[1 .5 0]*1,... 'MarkerEdgeColor',[1 .5 0]*1,'Markersize',4) plot(dp,0,'*k','Markersize',3) % Center of triangle % spx = (msx1 + msx2 + msx3)/3; % spy = (msy1 + msy2 + msy3)/3; % plot(spx,spy,'dg','MarkerFaceColor','g') % center of triangle % Scale force-space 20x plot([-18 2],[18 18],'k','LineWidth',2) text(-9,18.5,'100 % BW','VerticalAlignment','bottom',... 'HorizontalAlignment','center') text(-9,17.5,'20 mm','VerticalAlignment','top',... 'HorizontalAlignment','center') end % plot ground reactio force vectors for kkk = 5:10:95 quiver(pxm(kkk),pym(kkk),GRF(kkk,1)*20,GRF(kkk,2)*20,... 'color',cl_kk(kk,:),'LineWidth',1,... 'MaxHeadSize',.5,'AutoScaleFactor',1); end xlabel('x (mm)') ylabel('y (mm)') axis([-22 25 -22 25]) axis square %%%%%%%%%%%%%%%%%%%%%%%%%% % Sagittal view xz-plane % %%%%%%%%%%%%%%%%%%%%%%%%%% subplot(2,1,1) % Body height clear s s = scatter(zeros(length(BodyHeight),1),BodyHeight,... 'filled','SizeData',15,... 'markerfacecolor',[1 .5 0]*.8); alpha(s,.5) % Ground reaction force vectors for kkk = 5:10:95 quiver(pxm(kkk),0,GRF(kkk,1)*20,GRF(kkk,3)*20,... 'color',cl_kk(kk,:),'LineWidth',1,... 'MaxHeadSize',.5,'AutoScaleFactor',1); end % Thorax axis (Petiole Head-Thorax-joint) % plot layer as last layer if kk == 3 plot([b3px b2px],[b3pz b2pz],'color',[1 .5 0]*1,'linewidth',2) % Scale force-space 20x plot([-18 2],[18 18],'k','LineWidth',2) text(-9,18.5,'100 % BW','VerticalAlignment','bottom',... 'HorizontalAlignment','center') text(-9,17.5,'20 mm','VerticalAlignment','top',... 'HorizontalAlignment','center') end xlabel('x (mm)') ylabel('z (mm)') axis([-22 25 -22 25]) axis square end end
Fig. 3A-C: GRF over time

close all %figure(1) figure('Name','test', 'Units', 'normalized', 'Position', [0,0,1,1]) ha = tight_subplot(9,5,[0.01 0.01]*2,[0.03 0.01]*2,[0.03 0.03]*2); % constant of free fall g = 9.81; % [kg*m/s^2] clear slopes slopes = unique(data1.deg); % range green stripes ay1 = [1 1 1]*-.75; ay2 = [1 1 1]*.75; % color double support durations dscol = [0 .7 0]; % color leg cl_kk = [0 0 1; 0 .7 0; 1 0 0]; for k = 1:length(slopes) % slopes % filter for subsets clear filter_k filter_k = data1.deg==slopes(k); % Gait cycle duration time = data1.GaitCycleDuration(filter_k); % time in s % normalised stance duration per slope (middle legs, normalised) time_stance = data1.TimeStance2(filter_k)./time*100; % in percent % time_stance_m = mean(time_stance); for kk = 1:3 % legs filter_kk = data1.deg==slopes(k) & str2num(cell2mat(data1.legnr))==kk; for kkk = 1:3 % directions clear fq fqm fq1 fq2 % Force % Tripod 1 % F(degs,legs,tripod,direction,time,sample) fq = squeeze(F(k,kk,2,kkk,:,:)); fqm = mean(fq,2); % Tripod 2 fq2 = squeeze(F(k,kk,1,kkk,:,:)); fqm2 = mean(fq2,2); % plot ak = k + 15*kkk - 15 + 5*kk - 5; axes(ha(ak)); hold all box on fill([50 time_stance_m time_stance_m 50 50],... [ay1(kkk) ay1(kkk) ay2(kkk) ay2(kkk) ay1(kkk)],... [1 1 1]*.7,'EdgeColor',dscol,'FaceColor',dscol,... 'FaceAlpha',.2,'EdgeAlpha',0); hold all fill(100+[50 time_stance_m time_stance_m 50 50],... [ay1(kkk) ay1(kkk) ay2(kkk) ay2(kkk) ay1(kkk)],... [1 1 1]*.7,'EdgeColor',dscol,'FaceColor',dscol,... 'FaceAlpha',.2,'EdgeAlpha',0) fill([100 50+time_stance_m 50+time_stance_m 100 100],... [ay1(kkk) ay1(kkk) ay2(kkk) ay2(kkk) ay1(kkk)],... [1 1 1]*.7,'EdgeColor',dscol,'FaceColor',dscol,... 'FaceAlpha',.2,'EdgeAlpha',0) fill(-100+[100 50+time_stance_m 50+time_stance_m 100 100],... [ay1(kkk) ay1(kkk) ay2(kkk) ay2(kkk) ay1(kkk)],... [1 1 1]*.7,'EdgeColor',dscol,'FaceColor',dscol,... 'FaceAlpha',.2,'EdgeAlpha',0) % plot standard deviation stdshade(fq',1,'k',[1 1 1]*0.6,[1:length(fq)]); hold on % tripod 2 plot([1:length(fqm)],fqm2,'LineStyle','-','LineWidth',.4,... 'Color',[0 0 0]); hold on % tripod 1 plot([1:length(fqm)],fqm,'LineStyle','-','LineWidth',1.2,... 'Color',cl_kk(kk,:)); hold on % plot title only for first six subplots if ak < 6 title([num2str(slopes(k)),'°'],'FontWeight','normal', ... 'FontSize',10) end axis square end end end set(ha([1:5:45]),'YTickLabel',{'-0.5','0','0.5'},'YTick',[-0.5,0,0.5],'TickLength',[0.05,0.05]) set(ha(40:45),'XTickLabel',{'0','','100'},'XTick',[50,50 + time_stance_m,150],... 'XTickLabel',{'0','','100'},'XTick',[50,50 + time_stance_m,150],... 'TickLength',[0.05,0.05]) set(ha,'TickDir','in'); set(ha,'Xlim',[0 170],'Ylim',[-.75 .75]) xlabel(ha(43),'Stride cycle right middle leg (%)') ylabel(ha(6),'Fore-aft GRF (BW)','FontWeight','normal', 'FontSize',10,'linewidth',.5) ylabel(ha(21),'Lateral GRF (BW)','FontWeight','normal', 'FontSize',10,'linewidth',.5) ylabel(ha(36),'Normal GRF (BW)','FontWeight','normal', 'FontSize',10,'linewidth',.5) % label leg right axis set(ha([5:5:45]),'yaxislocation','right') yl = {'{\color{blue}L1} + R1', '{\color[rgb]{0,0.7,0}R2} + L2', '{\color[rgb]{1,0,0}L3} + R3',... '{\color{blue}L1} + R1', '{\color[rgb]{0,0.7,0}R2} + L2', '{\color[rgb]{1,0,0}L3} + R3',... '{\color{blue}L1} + R1', '{\color[rgb]{0,0.7,0}R2} + L2', '{\color[rgb]{1,0,0}L3} + R3',... }; ik = 0; for k = 5:5:45 ik = ik + 1; set(get(ha(k),'YLabel'),'String',cell2mat(yl(ik)),'FontSize',10,'FontWeight','normal','linewidth',.5); end
Fig. 4A-D: Comparison of the mean GRF vectors on upslopes and downslopes

close all % color leg pair cl_kk = [1 .7 0;0 0 1;0 .7 0]; % compare slopes deg_pair = [-60,60;-30,30]; % force-space ratio fscale = 20; % axis axis_vec=[-19 19 -19 19]; % midstance (mean of range between 49 to 51, [1:100]) meanwindow = 49:51; % scale settings force_scale_text = '50 % body weight'; space_scale_text = '10 mm'; scale = [-9,-9 + fscale*.5]; % changing linewidth for vectors lw = linspace(1,.1,105); for m = 1:2 % plot layers %(individual measurements below mean and below force) for l = 1:2 % slope pair clear degs degs = deg_pair(l,:); for k = 1:length(degs); % selected slope for kk = 1:3 % legs clear filter_kk filter_kk = data2.deg==degs(k) & data2.leg == num2str(kk); % 1 Tarsi % Anterior extreme positions aep % static supporting triangle clear aepx aepy aepz pet50x pet50y pet50z aepx = mean(data2.p5x(1:2,filter_kk))-mean(data2.b3x(1:2,filter_kk)); aepy = mean(data2.p5y(1:2,filter_kk))-mean(data2.b3y(1:2,filter_kk)); aepz = -mean(data2.b3z(1:2,filter_kk)); % 2 Location of Petiole during midstance % (relative to petiole location during touchdowns) pet50x = mean(data2.b3x(meanwindow,filter_kk))-mean(data2.b3x(1:2,filter_kk)); pet50y = mean(data2.b3y(meanwindow,filter_kk))-mean(data2.b3y(1:2,filter_kk)); pet50z = mean(data2.b3z(meanwindow,filter_kk))-mean(data2.b3z(1:2,filter_kk)); % ground reaction force clear fx fy fz fx = mean(data2.Cx(:,filter_kk),2)*fscale; fy = mean(data2.Cy(:,filter_kk),2)*fscale; fz = mean(data2.Cz(:,filter_kk),2)*fscale; % mirror lateral direction if leg == middle leg if kk == 2 pet50y=-pet50y; aepy = -aepy; fy = -fy; end % one figure per slope pair figure(l); % one subplot for each plane (xy versus xz) subplot(2,1,1) % Colors clear colo if (degs(k) < 0 && kk == 1) colo = [0 0 1]; elseif (degs(k) > 0 && kk == 1) colo = [0 0 0.3]; elseif (degs(k) < 0 && kk == 3) colo = [1 0 0]; elseif (degs(k) > 0 && kk == 3) colo = [0.3 0 0]; elseif (degs(k) < 0 && kk == 2) colo = [0 .7 0]; else colo = [0 .7 0]*.3; end % plot annotations and individual measurements % below mean and force if m == 1 % plot origin of coordiante system plot(0,0,'ok') % plot petiole mean+-std (crosses) plot(pet50x,pet50y,'.','color',[1 1 1].*0) % plot anterior extreme position plot(aepx,aepy,'+','color',[1 1 1]*.5) else plot(mean(aepx)+std(aepx)*[1 -1],mean(aepy).*[1 1],'k','Linewidth',1.5) plot(mean(aepx)*[1 1] ,mean(aepy)+std(aepy)*[1 -1],'k','Linewidth',1.5) % plot force for kkk = 1:5:length(fx) quiver(mean(aepx),mean(aepy),fx(kkk),fy(kkk),'Color',colo,'MaxHeadSize',.8,'AutoScaleFactor',1,... 'Linewidth',lw(kkk)); hold on end xlabel('x (mm)') ylabel('y (mm)') axis(axis_vec) axis square end % xz-plane subplot(2,1,2) % plot annotations and individual measurements % below mean and force if m == 1 % plot origin of coordinate system plot(0,0,'ok') % plot scale text(-9+(scale(2)-scale(1))/2,-9,force_scale_text,... 'FontSize',10,'VerticalAlignment','top',... 'HorizontalAlignment','center'); hold on text(-9+(scale(2)-scale(1))/2,-9,space_scale_text,... 'FontSize',10,'VerticalAlignment','bottom',... 'HorizontalAlignment','center'); hold on plot(scale,[-9, -9],'k','LineWidth',0.5) % plot petiole mean+-std (crosses) plot(pet50x,pet50z,'.','color',[1 1 1]*0) % plot anterior extreme position plot(aepx,aepz,'+','color',[1 1 1].*.5) else % plot mean anterior extreme position plot(mean(aepx)+std(aepx)*[1 -1],mean(aepz)*[1 1],'k','Linewidth',1.5) plot(mean(aepx)*[1 1] ,mean(aepz)+std(aepz)*[1 -1],'k','Linewidth',1.5) % plot force for kkk = 1:5:length(fx) clear ah quiver(mean(aepx),mean(aepz),fx(kkk),fz(kkk),'Color',colo,'MaxHeadSize',.8,... 'AutoScaleFactor',1,'Linewidth',lw(kkk)); hold on end xlabel('x (mm)') ylabel('z (mm)') axis(axis_vec) axis square end end end end end
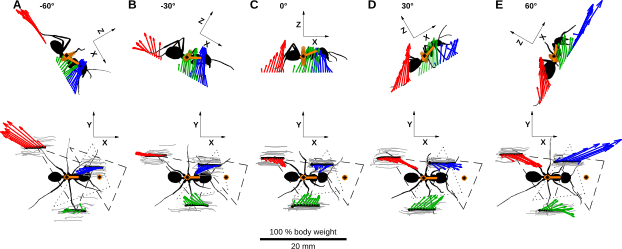
Fig. 5A-B: Hind leg dragging and change of body height
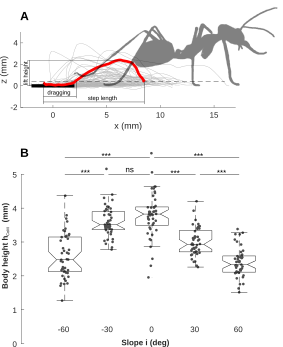
Fig. 5A: Hind leg dragging
close all % sample trajectory in red sam = 36; % Uncertainty xmin = 0.4; % [mm] % Hind leg Tarsus trajectories x = p5l.x; % zero vertical component z = p5l.z - repmat(mean(p5l.z(1:2,:)),100,1); % Plot % trajectories plot(x,z,'color',[1 1 1]*.7,'LineWidth',.5); hold on % uncertainty line plot([-2 16], [xmin xmin],'--k') % force plate plot([-2 2], [0 0],'k','Linewidth',5) % sample trajectory in red plot(x(:,sam),z(:,sam),'r','Linewidth',4); hold all axis equal axis([-3 17 -2 5 ]) xlabel('x (mm)') ylabel('z (mm)') shg
Fig. 5B: Change of body height
close all; clear bh % create dataframe for bodyheight bh.BodyHeight = data2.BodyHeight'; bh.leg = num2str(data2.leg); bh.deg = data2.deg; clear slopes slopes = unique(bh.deg); figure(1) for k = 1:5 % plane % subset data clear filter_k dat_k lx x2 filter_k = logical(bh.deg == slopes(k)); dat_k = bh.BodyHeight(filter_k); % scatter and plot data lx = length(dat_k); x2 = linspace(-.1,.1,lx); plot(x2+k,dat_k,'.','Color',[1 1 1]*.3,'Markersize',10); hold on end boxplot(bh.BodyHeight,bh.deg,... 'labelverbosity','majorminor',... 'medianstyle','line','notch','on',... 'boxstyle','outline','factordirection','list',... 'symbol','+k','outliersize',0.1,'widths',.75); xlabel('Slope i (deg)') ylabel('Body height (mm)')
Fig. 6A-C: Normalised double support durations for each leg pair at different slopes
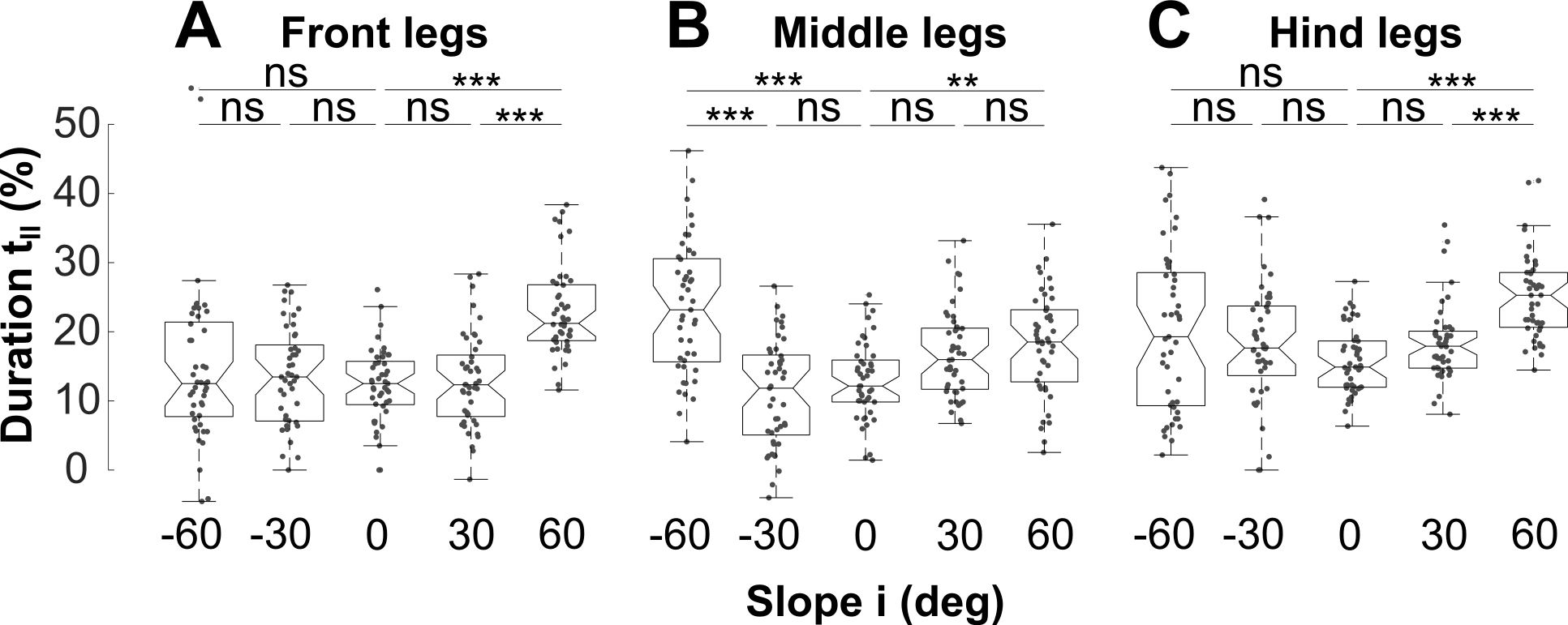
close all % stride duration middle leg per definition gait cycle duration clear sd slopes sd = data1.td22 - data1.td12; slopes = unique(data1.deg); for k = 1:3 % legs clear dat % normalise double support durations % by stride duration of the middle legs dat = eval(['data1.ds',num2str(k)])./sd*100; figure(k) for kk = 1:length(slopes) % slopes % subset data filter_kk = data1.deg == slopes(kk); dat_kk = dat(filter_kk); % scatter and plot data lx = length(dat_kk); x2 = linspace(-.1,.1,lx); plot(x2+kk,dat_kk,'.','Color',[1 1 1]*.3,'Markersize',10); hold on end % plot boxplot over scatterd data bh = boxplot(data1.(['ds',num2str(k)])./sd*100,data1.deg,... 'labelverbosity','majorminor',... 'medianstyle','line','notch','on',... 'boxstyle','outline','factordirection','list',... 'symbol','.','outliersize',0.1,'widths',.75,'color',[0 0 0]); xlabel('Slope i (deg)') ylabel('Duration (%)') end
stepx: stepping length in mm dx: dragging length in mm dz: tarsus lift heights in mm
grpstats(d3,'deg','median','DataVars',{'stepx','dx','dz'})
ans =
deg GroupCount median_stepx median_dx median_dz
-60 -60 15 6.8024 6.8264 0.42321
-30 -30 15 7.7551 7.767 0.774
0 0 15 9.1626 9.1626 0.996
30 30 15 8.574 8.574 1.0428
60 60 15 8.4687 8.4687 1.443
Table 4: Kinematic parameter
data1.Duty2 = data1.TimeStance2./data1.TimeStep2*100; % Duty factor data1.TimeStep2 = data1.TimeStep2.*1000; % Stride duration of the middle legs in ms data1.TimeStance2 = data1.TimeStance2.*1000; % Contact duration of the middle legs in ms data1.GaitCycleVelocity2 = data1.GaitCycleVelocity2.*1000; % Running speed in mm/s % Displacement of the CoM per stride of the middle legs in mm/Stride data1.GaitCycleDisplacementS2 = data1.GaitCycleDisplacementS2.*1000; grpstats(data1,'deg',{'mean','std'},'DataVars',... {'TimeStep2','TimeStance2','Duty2','GaitCycleVelocity2','GaitCycleDisplacementS2'})
ans =
deg GroupCount mean_TimeStep2 std_TimeStep2
-60 -60 45 127.87 29.746
-30 -30 45 108.98 14.082
0 0 45 109.91 12.634
30 30 45 135.87 22.504
60 60 45 168.98 19.751
mean_TimeStance2 std_TimeStance2 mean_Duty2 std_Duty2
-60 93.067 29.362 71.69 7.2404
-30 68.622 15.391 62.429 7.8164
0 68.933 10.75 62.681 6.1896
30 89.778 19.974 65.672 5.6179
60 114.44 20.23 67.483 6.8958
mean_GaitCycleVelocity2 std_GaitCycleVelocity2
-60 63.777 25.275
-30 83.723 26.113
0 87.362 17.402
30 70.642 14.593
60 53.673 9.6925
mean_GaitCycleDisplacementS2 std_GaitCycleDisplacementS2
-60 7.7211 1.9294
-30 9.0467 2.0994
0 9.6601 1.5619
30 9.4821 1.1748
60 9.0547 1.2206
Table 4: Body Height (Center of mass - substrate distance in mm)
data3.deg = data2.deg; data3.BodyHeight = data2.BodyHeight'; grpstats(dataset(data3),'deg',{'mean','std'},'DataVars','BodyHeight')
ans =
deg GroupCount mean_BodyHeight std_BodyHeight
-60 -60 45 2.582 0.6612
-30 -30 45 3.596 0.45271
0 0 45 3.7922 0.67355
30 30 45 3.0065 0.41909
60 60 45 2.399 0.43189
Body Height (Petiole - substrate distance in mm)
grpstats(data1,'deg',{'mean','std'},'DataVars','BodyHeightMean')
ans =
deg GroupCount mean_BodyHeightMean std_BodyHeightMean
-60 -60 45 2.1749 0.61942
-30 -30 45 3.2094 0.45431
0 0 45 3.4913 0.65939
30 30 45 2.7155 0.40139
60 60 45 2.116 0.41245